AMBER#
Versions Installed
Kay: 16 / 18
LXP: 22
Description#
AMBER (Assisted Model Building with Energy Refinement)[1], is a general purpose molecular mechanics/dynamics suite which uses analytic potential energy functions, derived from experimental and ab initio data, to refine macromolecular conformations.
License#
ICHEC has acquired a site license for the Amber 16 (AmberTools 17) and Amber 18 (AmberTools 18) packages.
Job Submission example#
A job submission example is as follows.
On Kay#
#!/bin/bash -l
# All the informations about queues can be obtained by 'sinfo'
# PARTITION AVAIL TIMELIMIT
# DevQ up 1:00:00
# ProdQ* up 3-00:00:00
# LongQ up 6-00:00:00
# ShmemQ up 3-00:00:00
# PhiQ up 1-00:00:00
# GpuQ up 2-00:00:00
# Slurm flags
#SBATCH -p ProdQ
#SBATCH -N 2
#SBATCH --job-name=jobname
#SBATCH -t 60:00:00
# Charge job to myaccount
#SBATCH -A your_project
# Write stdout+stderr to file
#SBATCH -o output.txt
# Mail me on job start & end
#SBATCH --mail-user=your_email
#SBATCH --mail-type=BEGIN,END
cd $SLURM_SUBMIT_DIR
module load amber/16
module load intel/2019
module load gcc/8.2.0
srun $AMBERHOME/bin/pmemd.MPI -O -i mdin.in -o mdout.out -p prmtop.prm -c inpcrd.rst -ref inpcrd.rst -x trjfile.trj -inf file.info -r file.rst7
On Meluxina#
#!/bin/bash -l
# Slurm flags
#SBATCH -p cpu
#SBATCH --qos default
#SBATCH -N 2
#SBATCH --job-name=jobname
#SBATCH --hint=nomultithread
#SBATCH -t 60:00:00
# Charge job to myaccount
#SBATCH -A your_project
# Write stdout+stderr to file
#SBATCH -o output.txt
# Mail me on job start & end
#SBATCH --mail-user=your_email
#SBATCH --mail-type=BEGIN,END
cd $SLURM_SUBMIT_DIR
module load amber/22-foss-2023a-CUDA-12.2
srun $AMBERHOME/bin/pmemd.MPI -O -i mdin.in -o mdout.out -p prmtop.prm -c inpcrd.rst -ref inpcrd.rst -x trjfile.trj -inf file.info -r file.rst7
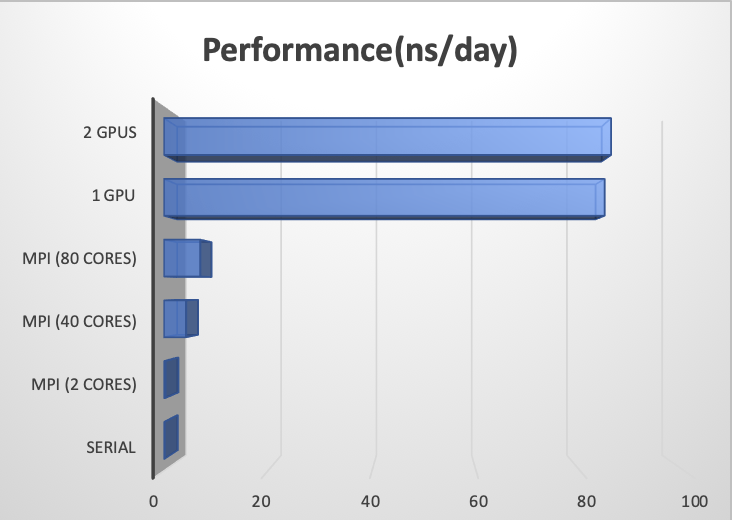
Benchmarks#
Version: Amber 18
Dataset: Cellulose (408,609 atoms) NPT Ensemble
Merit: ns/day (the higher the better)
Resource |
Performance(ns/day) |
|---|---|
Serial |
0.15 |
MPI (2 cores) |
0.28 |
MPI (40 cores) |
4.27 |
MPI (80 cores) |
6.99 |
1 GPU |
84.71 |
2 GPUs |
85.94 |
Clearly, the speedup is enormous on GPU compared to CPUs.
xychart-beta
title "Performace vs Resource"
x-axis ["Serial", "MPI (2 cores)", "MPI (40 cores)", "MPI (80 cores)", "1 GPU", "2 GPUs"]
y-axis "Performace (ns/day)" 0 --> 90
bar [0.15, 0.28, 4.27, 6.99, 84.71, 85.94]
Additional Notes#
To use Amber 18 load the relevant environment module: module load amber/18. Use pmemd.MPI for MPI-only version, pmemd.cuda for CUDA-only version, pmemd.cuda.MPI for CUDA+MPI version.
